Fraud detection
Understanding the data
df = pd.read_csv('creditcard.csv')
df.head()
| Time | Amount | class | v1 | v28 |
|---|---|---|---|---|
| 1.0 | 149.62 | 0 | -1.359807 | -0.021053 |
- NOTE There are 28 ‘v’ features : v1,…,v28
- Due to privacy reasons most colums dont have a column name
- Time : not enough information about this feature, going to remove
- amount : the monentary amount of the transaction that was flagged as fraud
- classes :
- 0: No fraud
- 1: Fraud
Exploratory Analysis (EDA)
plt.bar(x=[0,1],height=[df['Class'].value_counts()[0],df['Class'].value_counts()[1]])
plt.title('Class Distributions (0:No Fraud 1: Fraud)', fontsize=14)
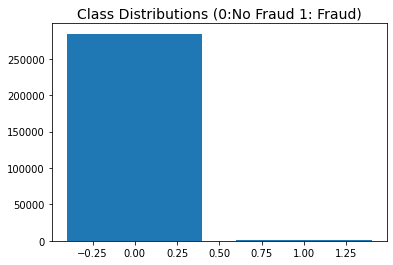
| Class | |
|---|---|
| 0 | 284315 |
| 1 | 492 |
amount_val = df['Amount'].values
fig,ax = plt.subplots()
sns.distplot(amount_val, color='r',ax=ax)
ax.set(title='Distribution of Transaction Amount')
ax.set_xlim([min(amount_val), max(amount_val)])
plt.show()

- Tansactions are mainly of smaller amounts
Metric Selection
- very imbalanced class distribution, positive class is minority
- positive class more important than negative class, need to detect if fraud occurs with high confidence
- e.g. 90% precision
- False negatives; falsely classifiying fraud sample as not fraud is very costly need to minimise.
- need as low as possible
- trade-off: high recall as possible due to False negatives
- but precision needs to be above a threshold to be useful
- Precision, recall,f1 measure and False negatives going to be used to compare models via cv
Pre-processing
- Dataset was scaled, used robust scaler which is more robust to outliers
- To better visualise the data and conduct multi-vartiate analysis, will implement a random under sampling technique, which undersamples the majority class, creating a more balanced dataset
rob_scaler = RobustScaler()
X = df.drop(['Class','Time'],axis=1)
y = df['Class']
X_scaled = rob_scaler.fit_transform(X)
X_scaled_df = pd.DataFrame(X_scaled, columns = X.columns)
rus = RandomUnderSampler(random_state=42)
X, y = rus.fit_resample(X, y)
sns.countplot(y)

balanced_df = pd.DataFrame(X.join(y))
corr = balanced_df.corr()
sns.heatmap(corr, cmap='coolwarm_r', annot_kws={'size':20}).set_title("Imbalanced Correlation Matrix", fontsize=14)
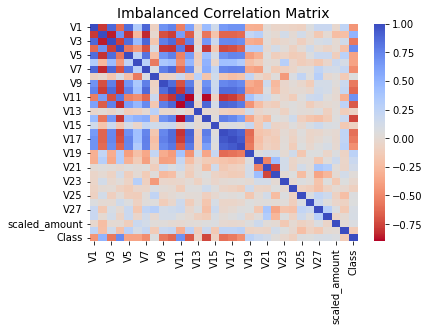
- v4, v11 features are highly positively correlated with class
- v14 andv10 are highly negatively skewed with class
- no features are hightly correlated with eachother
- if there was highly correlated features , should remove
Model selection
- Needed to split the data into training and test sets, then apply transformation in pipeline
- to prevent leakage when estimating model preformance on via cross-validation.
- Used stratified random sampling, as dataset very imbalanced
- reloaded orginal unbalanced dataset
X = df.drop(['Class','Time'],axis=1)
y= df['Class']
X_train, X_test, y_train, y_test = train_test_split(X,y,stratify=y,random_state=1)
Baseline score, without resampling or specialised imbalanced algorithms
- First going to use:
- naive model: dummy classifier which outputs uniform results
- logistic regression
- random forest
- neural network
- Aim: get a baseline score without specialised algorithms designed for imbalanced learning
- naive algorithms without resampling etc
- The first 3 models were implemented using the sci-kitlearn library
- stratified 10 fold cv was used to calculate f1, precision and recall values
- The neural network model was implemented using the tensorflow library
sci-kit learn (model 1,2 and 3)
from sklearn.dummy import DummyClassifier
from sklearn.pipeline import Pipeline
pipe1 = Pipeline([
('scaler',RobustScaler()),
('model',DummyClassifier(strategy='uniform'))
])
pipe2 = Pipeline([
('scaler',RobustScaler()),
('model',LogisticRegression())
])
pipe3 = Pipeline([
('scaler',RobustScaler()),
('model',RandomForestClassifier())
])
from sklearn.model_selection import cross_validate
from sklearn.model_selection import StratifiedKFold
cv = StratifiedKFold(n_splits=10)
scores= ['f1','precision','recall']
navie_cv_results = cross_validate(pipe1,X=X_train,y=y_train,cv=cv,scoring=scores,n_jobs=-1)
logistic_cv_results = cross_validate(pipe2,X=X_train,y=y_train,cv=cv,scoring=scores,n_jobs=-1)
randomForest_cv_results = cross_validate(pipe3,X=X_train,y=y_train,cv=cv,scoring=scores,n_jobs=-1)
The mean f1, precision and recall scores
model_1: naive algo
| 0 | |
|---|---|
| test_f1 | 0.00347783 |
| test_precision | 0.00174494 |
| test_recall | 0.503979 |
model_2: logistic regression
| 0 | |
|---|---|
| test_f1 | 0.685409 |
| test_precision | 0.852982 |
| test_recall | 0.577477 |
model_3: random forest
| 0 | |
|---|---|
| test_f1 | 0.840989 |
| test_precision | 0.948857 |
| test_recall | 0.756156 |
- clearly the naive algo performs the worst
- mean scores from cv does not show the whole picture
- must visualise learning curves to diagnose any problems in the learning stage
The precision and recall learning curves
model_2: logistic regression
_algo_learning_curve.png)
- logistic regression seems to be the best, but recall score is low
model_3: random forest
_learning_curves.png)
- randomforest is overfitting
- represented by large gap between the training and test scores
Tensorflow neural network
- The neural network will be built via a different library called keras
METRICS = [
keras.metrics.Precision
keras.metrics.FalseNegatives
keras.metrics.Recall
]
def make_model(metrics=METRICS, output_bias=None):
if output_bias is not None:
output_bias = tf.keras.initializers.Constant(output_bias)
model = keras.Sequential([
keras.layers.Dense(
16, activation='relu',
input_shape=(train_features.shape[-1],)),
keras.layers.Dropout(0.5),
keras.layers.Dense(1, activation='sigmoid',
bias_initializer=output_bias),
])
model.compile(
optimizer=keras.optimizers.Adam(learning_rate=1e-3),
loss=keras.losses.BinaryCrossentropy(),
metrics=metrics)
return model
model.compile(
optimizer=keras.optimizers.Adam(learning_rate=1e-3),
loss=keras.losses.BinaryCrossentropy(),
metrics=metrics)
return model
EPOCHS = 100
BATCH_SIZE = 2048
early_stopping = tf.keras.callbacks.EarlyStopping(
monitor='val_prc',
verbose=1,
patience=10,
mode='max',
restore_best_weights=True)
model = make_model()
model.summary()
| type | Output Shape | Param |
|---|---|---|
| Dense | (None, 16) | 480 |
| Dropout | (None, 16) | 0 |
| Dense | (None, 1) | 17 |
- Total params: 497
- Trainable params: 497
- Non-trainable params: 0
_learning_curve.png)
concluding first stage
- logistic regression and Random Forest is shortlisted
- the complexity of the algorithms will be increased
Specialised algorithms
-
Implemented SMOTE and cost-sensitive learning on the shortlisted model
-
logic of using cv for imbalanced learning:
- up/down sampling done in inside cv
- imblearn.pipeline extends scikit-learn pipeline.
- Up/downsampled only the data in the training section
- Fit the model on the up/down sampled training data
- Score the model on the (non-up/downsampled) validation data
recall/precision trade-off
- SMOTE and balanced class wieghts increases recall at the cost of precision
from imblearn.pipeline import Pipeline as imbpipeline
pipeline1 = imbpipeline([
['smote', SMOTE(random_state=11)],
['scaler',RobustScaler()],
['classifier', LogisticRegression(random_state=11,penalty='l2', n_jobs=-1, max_iter=1000)]
])
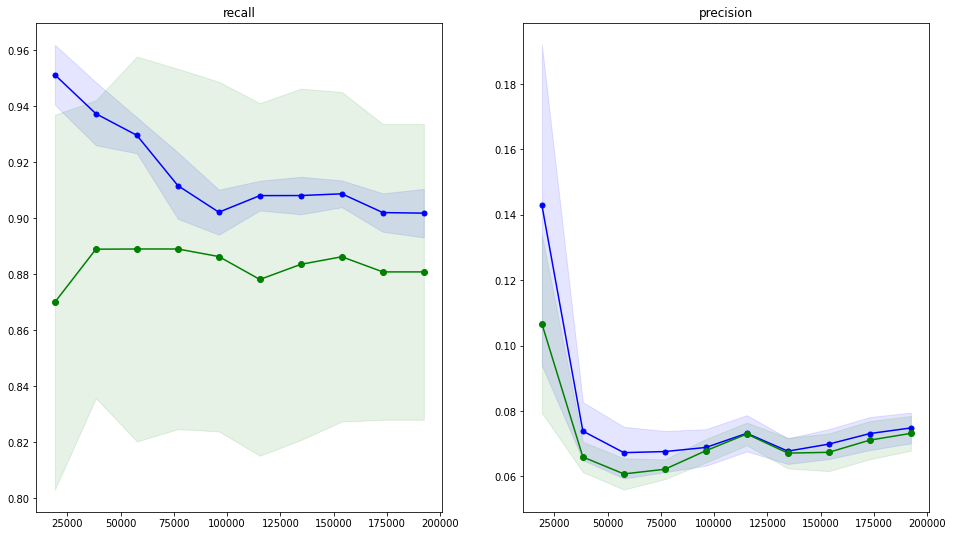
- precision of ~0.03 is not acceptable